Abstract
Research Article
Characterization of plastic degrading bacteria isolated from landfill sites
Afreen Bakht, Nouman Rasool* and Saima Iftikhar
Published: 28 May, 2020 | Volume 3 - Issue 1 | Pages: 030-035
The plastic pollution is threatening the environment because it has very slow degradation rate and high usage in regular activities. The present study aims at the isolation of novel microorganisms that would assist faster degradation process of polyethylene. The waste samples were collected from different landfills and dumpsites. Out of forty samples, eight samples were found to degrade polythene strips in liquid medium. Further screening of these samples showed that two strains of microbes had high potential for polythene degradation. Biochemical tests and ribotyping were performed for characterization of isolated bacteria. Resultantly, two novel bacterial strains were identified named; Bacillus wudalianchiensis_UMT (2A) and Pseudomonas aeruginosa_UMT (6). Analysis of these microbes further revealed that Bacillus wudalianchiensis_UMT and Pseudomonas aeruginosa_UMT have capability to degrade 6.6% and 4.8% polyethylene respectively. So, the results disclosed that these bacteria have great potential to degrade polythene in less time as compare to natural degradation process and can contribute to reduce pollution from our environment.
Read Full Article HTML DOI: 10.29328/journal.ijcmbt.1001013 Cite this Article Read Full Article PDF
References
- Kint D, Muñoz‐Guerra S. A review on the potential biodegradability of poly (ethylene terephthalate). Polymer Int. 1999; 48: 346-352. PubMed:
- Shaw DK, Sahni P. Plastic to oil. J Mechan Civil Engineering. 2004; 46-48.
- Zhang J, Wang X, Gong J, Gu Z. A study on the biodegradability of polyethylene terephthalate fiber and diethylene glycol terephthalate. J Applied Polymer Sci. 2004; 93: 1089-1096.
- Tansel B, Yildiz BS. Goal-based waste management strategy to reduce persistence of contaminants in leachate at municipal solid waste landfills. Environment, Development and Sustainability. 2001; 13: 821-831.
- Sinha V, Patel MR, Patel JV. PET waste management by chemical recycling: a review. J Polymers Environ. 2010; 18: 8-25.
- Astrup T, Møller J, Fruergaard T. Incineration and co-combustion of waste: accounting of greenhouse gases and global warming contributions. Waste Manag Res. 2009; 27: 789-799. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/19748939
- Yamada-Onodera K, Mukumoto H, Katsuyaya Y, Saiganji A, Tani Y. Degradation of polyethylene by a fungus, Penicilliumsimplicissimum YK. Polymer Degradation and Stability. 2001; 72: 323-327.
- Head IM, Swannell, RP. Bioremediation of petroleum hydrocarbon contaminants in marine habitats. Curr Opin Biotechnol. 1999; 10: 234-239. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/10361073
- Santhini K, Myla J, Sajani S, Usharani G. Screening of Micrococcus sp from oil contaminated soil with reference to bioremediation. Botany Res Int. 2009; 2: 248-252.
- Yoon MG, Jeon HJ, Kim MN. Biodegradation of polyethylene by a soil bacterium and AlkB cloned recombinant cell. J Bioremed Biodegrad. 2012; 3: 1-8.
- Kim MN, Park ST. Degradation of poly (L‐lactide) by a mesophilic bacterium. J Appl Polymer Sci. 2010; 117: 67-74.
- Augusta J, Müller RJ, Widdecke HA. A rapid evaluation plate-test for the biodegradability of plastics. Appl Microbiol Biotechnol. 1993; 39: 673-678.
- Usha R, Sangeetha T, Palaniswamy M. Screening of polyethylene degrading microorganisms from garbage soil. Libyan Agri Res Center J Int. 2011; 2: 200-204.
- Kwon SW, Kim JS, Park IC, Yoon SH, Park DH, et al. Pseudomonas koreensis sp. nov, Pseudomonas umsongensis sp. nov. and Pseudomonas jinjuensis sp. nov, novel species from farm soils in Korea. Int J Syst Evol Microbiol. 2003; 53: 21-27. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/12656147
- Rasool N, Rashid N, Javed MA, Haider MS. Requirement of pro-peptide in proper folding of subtilisin-like serine protease TK0076. Pak J Bot. 2011; 43: 2059-2065.
- Kapri A, Zaidi MGH, Satlewal A, Goel R. SPION-accelerated biodegradation of low-density polyethylene by indigenous microbial consortium. Int Biodeterio Biodegrad. 2010; 64: 238-244.
- Jalal A, Rashid N, Rasool N, Akhtar M. Gene cloning and characterization of a xylanase from a newly isolated Bacillus subtilis strain R5. J Bioscience Bioengineering. 2009; 107: 360-365.
- Vignesh R, Deepika RC, Manigandan P, Janani R. Screening ofdegrading microbes from various dumped soil samples. Int Res J Engineering Technol. 2016; 4: 2493-2498.
- Ribón W. Biochemical isolation and identification of mycobacteria. In Biochemical Testing. InTech. 2012.
- Brown A, Smith H. Benson's Microbiological Applications, Laboratory Manual in General Microbiology, Short Version. McGraw-Hill Science/Engineering/Math. 2014.
- Del Valle LJ, Jaramillo ML, Talledo M, Pons MJ, Flores L, et al. Development of a 16S rRNA PCR-RFLP assay for Bartonella identification: applicability in the identification of species involved in human infections. Uni J Microbiol Res. 2014.
- Munna MS, Tahera J, Afrad MMH, Nur IT, Noor R. Survival of Bacillus Spp. SUBB01 at high temperatures and a preliminary assessment of its ability to protect heat-stressed Escherichia coli cells. BMC Res Notes. 2015; 8: 637. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/26526722
- Amako K, Takade A. The fine structure of Bacillus subtilis revealed by the rapid-freezing and substitution-fixation method. Microscopy. J Electron Microsc (Tokyo). 1985; 34: 13-17. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/3932576
Figures:
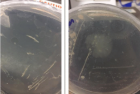
Figure 1
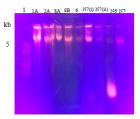
Figure 2
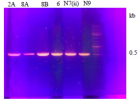
Figure 3

Figure 4

Figure 5

Figure 6

Figure 7

Figure 8
Similar Articles
-
Oral Candida colonization in HIV-infected patients: Species and antifungal susceptibility in Tripoli/LibyaEllabib M*,Mohamed H,Mokthar E,Ellabib M,El Magrahi H,Eshwika A. Oral Candida colonization in HIV-infected patients: Species and antifungal susceptibility in Tripoli/Libya . . 2018 doi: 10.29328/journal.ijcmbt.1001001; 1: 001-008
-
Evaluation of novel culture media prepared from plant substrates for isolation and identification of Cryptococcus Neoformans Species ComplexEllabib M*,Krema ZA,Mokthar ES,El Magrahi HS,Eshwika A,Cogliati M. Evaluation of novel culture media prepared from plant substrates for isolation and identification of Cryptococcus Neoformans Species Complex. . 2018 doi: 10.29328/journal.ijcmbt.1001002; 1: 009-013
-
Trypanosoma dionisii as an experimental model to study anti-Trypanosoma cruzi drugs: A comparative analysis with benznidazole, posaconazole and amiodaroneDe Souza W*,Barrias ES,Borges TR. Trypanosoma dionisii as an experimental model to study anti-Trypanosoma cruzi drugs: A comparative analysis with benznidazole, posaconazole and amiodarone. . 2018 doi: 10.29328/journal.ijcmbt.1001003; 1: 014-023
-
A Review on filaricidal activity of phytochemical extracts against filariasis and the Parasites Genomic DiversityAM Gumel*,MM Dogara. A Review on filaricidal activity of phytochemical extracts against filariasis and the Parasites Genomic Diversity. . 2018 doi: 10.29328/journal.ijcmbt.1001004; 1: 024-032
-
Host biomarkers for early diagnosis of infectious diseases: A comprehensive reviewArindam Chakraborty*,Singh Monica. Host biomarkers for early diagnosis of infectious diseases: A comprehensive review. . 2019 doi: 10.29328/journal.ijcmbt.1001005; 2: 001-007
-
Virulence Genes in Pseudomonas Aeruginosa Strains Isolated at Suez Canal University Hospitals with Respect to the Site of Infection and Antimicrobial ResistanceNermine Elmaraghy*,Said Abbadi,Gehan Elhadidi,Asmaa Hashem,Asmaa Yousef. Virulence Genes in Pseudomonas Aeruginosa Strains Isolated at Suez Canal University Hospitals with Respect to the Site of Infection and Antimicrobial Resistance. . 2019 doi: 10.29328/journal.ijcmbt.1001006; 2: 008-019
-
Knowledge, perception and practices of Suez Canal University students regarding Hepatitis C Virus infection risk and means of preventionNermine Elmaraghy*,Hesham El-Sayed,Sohair Mehanna,Adel Hassan,Mahmoud Sheded,Maha Abdel-Fattah,Samar Elfiky,Nehal Lotfy,Zeinab Khadr. Knowledge, perception and practices of Suez Canal University students regarding Hepatitis C Virus infection risk and means of prevention. . 2019 doi: 10.29328/journal.ijcmbt.1001007; 2: 020-027
-
in silico discovery of potential inhibitors against Dipeptidyl Peptidase-4: A major biological target of Type-2 diabetes mellitusNouman Rasool*,Andleeb Subhani,Waqar Hussain,Nadia Arif. in silico discovery of potential inhibitors against Dipeptidyl Peptidase-4: A major biological target of Type-2 diabetes mellitus. . 2020 doi: 10.29328/journal.ijcmbt.1001008; 3: 001-010
-
Production and evaluation of enzyme-modified lighvan cheese using different levels of commercial enzymesMohammad B Habibi Najafi*,Mohammad Amin Miri. Production and evaluation of enzyme-modified lighvan cheese using different levels of commercial enzymes. . 2020 doi: 10.29328/journal.ijcmbt.1001009; 3: 011-016
-
Development of ELISA based detection system against C. botulinum type BArti Sharma*,S Ponmariappan. Development of ELISA based detection system against C. botulinum type B. . 2020 doi: 10.29328/journal.ijcmbt.1001010; 3: 017-020
Recently Viewed
-
Febrile Lumbar Pain Revealing a Massive Collection: Complicated Psoas Abscess Managed SurgicallyMohammed Amine Elafari*,Mamad Ayoub,Mohammed Amine Bibat,Rhayour Anas,Maachi Youssef,Amine Slaoui,Tarik Karmouni,Abdelatif Koutani,Khalid Elkhader. Febrile Lumbar Pain Revealing a Massive Collection: Complicated Psoas Abscess Managed Surgically. J Clin Med Exp Images. 2026: doi: 10.29328/journal.jcmei.1001041; 10: 010-012
-
Impact of Microplastics on Human Health through the Consumption of Seafood: A ReviewNeeraj Kumar*,Dev Brat Mishra. Impact of Microplastics on Human Health through the Consumption of Seafood: A Review. J Clin Med Exp Images. 2025: doi: 10.29328/journal.jcmei.1001036; 9: 015-019
-
Phenotypic differences in Obese Patients with Heart Failure with Preserved Ejection Fraction (HFpEF) - A Mini ReviewMichelle Nanni*, Vivian Hu, Swagata Patnaik, Alejandro Folch Sandoval, Johanna Contreras. Phenotypic differences in Obese Patients with Heart Failure with Preserved Ejection Fraction (HFpEF) - A Mini Review. New Insights Obes Gene Beyond. 2024: doi: 10.29328/journal.niogb.1001020; 8: 001-005
-
Stone on the Mesh: Intravesical Erosion after Laparoscopic Promontofixation-A Hidden Cost of DurabilityAyoub Mamad*,Mohammed Amine Bibat,Mohammed Amine Elafari,Midaoui Moncef,Amine Slaoui,Tarik Karmouni,Abdelatif Koutani,Khalid Elkhader. Stone on the Mesh: Intravesical Erosion after Laparoscopic Promontofixation-A Hidden Cost of Durability. J Clin Med Exp Images. 2026: doi: 10.29328/journal.jcmei.1001040; 10: 006-009
-
Feasibility study on the evaluation of the effect of narrow-band CE-Chirp ASSR in the hearing field after hearing aid in hearing-impaired childrenWang Yonghua*,Xing Shuoyao. Feasibility study on the evaluation of the effect of narrow-band CE-Chirp ASSR in the hearing field after hearing aid in hearing-impaired children. Adv Treat ENT Disord. 2019: doi: 10.29328/journal.ated.1001007; 3: 007-011
Most Viewed
-
Effects of dietary supplementation on progression to type 2 diabetes in subjects with prediabetes: a single center randomized double-blind placebo-controlled trialSathit Niramitmahapanya*,Preeyapat Chattieng,Tiersidh Nasomphan,Korbtham Sathirakul. Effects of dietary supplementation on progression to type 2 diabetes in subjects with prediabetes: a single center randomized double-blind placebo-controlled trial. Ann Clin Endocrinol Metabol. 2023 doi: 10.29328/journal.acem.1001026; 7: 00-007
-
Physical Performance in the Overweight/Obesity Children Evaluation and RehabilitationCristina Popescu, Mircea-Sebastian Șerbănescu, Gigi Calin*, Magdalena Rodica Trăistaru. Physical Performance in the Overweight/Obesity Children Evaluation and Rehabilitation. Ann Clin Endocrinol Metabol. 2024 doi: 10.29328/journal.acem.1001030; 8: 004-012
-
Hypercalcaemic Crisis Associated with Hyperthyroidism: A Rare and Challenging PresentationKarthik Baburaj*, Priya Thottiyil Nair, Abeed Hussain, Vimal MV. Hypercalcaemic Crisis Associated with Hyperthyroidism: A Rare and Challenging Presentation. Ann Clin Endocrinol Metabol. 2024 doi: 10.29328/journal.acem.1001029; 8: 001-003
-
Exceptional cancer responders: A zone-to-goDaniel Gandia,Cecilia Suárez*. Exceptional cancer responders: A zone-to-go. Arch Cancer Sci Ther. 2023 doi: 10.29328/journal.acst.1001033; 7: 001-002
-
The benefits of biochemical bone markersSek Aksaranugraha*. The benefits of biochemical bone markers. Int J Bone Marrow Res. 2020 doi: 10.29328/journal.ijbmr.1001013; 3: 027-031

If you are already a member of our network and need to keep track of any developments regarding a question you have already submitted, click "take me to my Query."