Abstract
Research Article
Evaluation of novel culture media prepared from plant substrates for isolation and identification of Cryptococcus Neoformans Species Complex
Ellabib M*, Krema ZA, Mokthar ES, El Magrahi HS, Eshwika A and Cogliati M
Published: 14 August, 2018 | Volume 1 - Issue 1 | Pages: 009-013
Background: Melanin production due to phenoloxidase activity is a distinctive property of Cryptococcus neoformans and Cryptococcus gattii species complex yeasts. Therefore, an agar medium containing a precursor of melanin pigment is potentially useful to identify and differentiate cryptococcal colonies from other yeasts.
Background: This study aimed to evaluate and compare the ability of Cryptococcus neoformans species complex isolates to produce brown-pigmented colonies when grown on media prepared from various plant leaves or seeds extracts.
Material and Methods: Forty-six C. neoformans species complex isolates which were obtained from various environmental and clinical samples were inoculated on different media containing coriander, cumin, soybean, lupine, flax, pumpkin, basil, peppermint, and marjoram, were observed for the rate of growth and pigment production during a five-day period.
Results: All isolates were pigmented on all media within 24-48 hours, and brown or dark brown colonies were observed in less than five days, while C. albicans grew but did not produce any pigment.
Conclusion: The differential media tested in the present study are simple and inexpensive, and represent alternative valid tools for presumptive identification of C. neoformans species complex from clinical and environmental samples
Read Full Article HTML DOI: 10.29328/journal.ijcmbt.1001002 Cite this Article Read Full Article PDF
Keywords:
Cryptococcus neoformans; Melanin production; Differential medium; Seeds, Leaves
References
- Rajasingham R, Smith RM, Park BJ, Jarvis JN, Govender NP, et al. Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect Dis. 2017; 17: 873-881. Ref.: https://tinyurl.com/y9kkgk8e
- Heitman J, Kozel TR, Kwon-Chung KJ, Perfect JR, Casadevall A. Cryptococcus: From Human Pathogen to Model Yeast. ASM Press, Washington, DC, USA. 2011. Ref.: https://tinyurl.com/yd53oxge
- De Hoog GS, Guarro J, Gene J, Figueras MJ. Atlas of clinical fungi. ASM Press, Washington, DC, USA. 2000.
- Lacaz CS, Porto E, Martins JEC. Micologia media: fungos, actinomicetos ealgas de interesse medico. 8th ed. Sao Paulo: Sarvier. 1991.
- Chaskes S, Tyndall RL. Pigment production by Cryptococcus neoformans from para and ortho-diphenols: effect of the nitrogen source. J Clin Microbiol. 1975; 1: 509-514. Ref.: https://tinyurl.com/y8us93ja
- Pal M, Mehrotra BS. Studies on the efficacy of sunflower seed agar for the isolation and identification of Cryptococcus neoformans. Arogya J Health Sci 1982; 8: 74-79.
- Strachan AA, Yu RJ, Blank F. Pigment production of Cryptococcus neoformans grown with extracts of Guizotia abyssinica. Appl Microbiol. 1971; 22: 478-489.
- Hopfer RL, Blank F. Caffeic acid-containing medium for identification of Cryptococcus neoformans. J Clin Microbiol. 1976; 2: 115-120. Ref.: https://tinyurl.com/ycwcbdn4
- Nandhakumar B, Kumar G, Prabhu CP, Menon T. Mustard seed agar,a new medium for the differentiation of Cryptococccus neoformans. J. Clin. Microbiol. 2006; 44: 674. Ref.: https://tinyurl.com/y78ymenx
- Nandhakumar B, Menon T, Kumar G. A new henna-based medium for the differentiation of Cryptococcus neoformans. J Med Microbiol. 2007; 56: 568. Ref.: https://tinyurl.com/yba8zu4q
- Tendolkar U, Tainwala S, Jog S, Mathur M. Use of a new medium - tobacco agar, for pigment production of Cryptococcus neoformans. Indian J Med Microbiol. 2003; 21: 277-279. Ref.: https://tinyurl.com/ybtory4b
- Hamzia Ali Ajah. Two new media apple leaves agar and eggplant leaves agar for identification of Cryptococcus neoformans. J Biology, Agriculture and Healthcare. 2014; 4. Ref.: https://tinyurl.com/y7al3pva
- Ellabib MS, Krema ZA, Allafi AA, Cogliati M. First report of two cases of cryptococcosis in Tripoli, Libya, infected with Cryptococcus neoformans isolates present in the urban area. J Mycol Med. 2017; 27: 421-424. Ref.: https://tinyurl.com/ycwn9gb4
- Ellabib MS, Aboshkiwa MA, Husien WM, D'Amicis R, Cogliati M. Isolation, identification and molecular typing of Cryptococcus neoformans from pigeon droppings and other environmental sources in Tripoli, Libya. Mycopathologia. 2016; 181: 603-608. Ref.: https://tinyurl.com/ycbvydzl
- Springer DJ, Mohan R, Heitman J. Plants promote mating and dispersal of the human pathogenic fungus Cryptococcus. PLoS One. 2017; 12: e0171695. Ref.: https://tinyurl.com/y8sr4ctv
- Rajeshwari CU, Andallu B. Reverse phase HPLC for the detection of flavonoids in the ethanolic extract of Coriandrum sativum L seeds. International Journal of Basic and Applied Sciences. 2012; 1: 21-26.
- Claudia-CT, Neli-Kinga O, Laurian V, Cristina M. Comparative Studies on Polyphenolic Composition, Antioxidant and Diuretic Effects of Nigella sativa L. (Black Cumin) and Nigella damascena L. (Lady-in-a-Mist) Seeds. Molecules. 2015; 20: 9560-9574. Ref.: https://tinyurl.com/y88sgcq9
- Montserrat D, Teresa H, Sergio R, Grzegorz L, Isabel E, et al. Bioactive Phenolic Compounds of Soybean (Glycine max cv. Merit): Modifications by Different Microbiological Fermentations. Pol J Food Nutr Sci. 2012; 62: 241-250. Ref.: https://tinyurl.com/ya8azlua
- Aleksander S, Jaroslaw C, Piotr K, Krzysztof D, Eleonora LS, et al . Antioxidant activity and phenolic content in three lupin species. Journal of Food Composition and Analysis. 2012; 25: 190-197. Ref.: https://tinyurl.com/y7fvumzg
Figures:
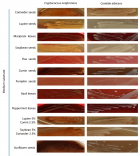
Figure 1
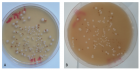
Figure 2
Similar Articles
-
Evaluation of novel culture media prepared from plant substrates for isolation and identification of Cryptococcus Neoformans Species ComplexEllabib M*,Krema ZA,Mokthar ES,El Magrahi HS,Eshwika A,Cogliati M. Evaluation of novel culture media prepared from plant substrates for isolation and identification of Cryptococcus Neoformans Species Complex. . 2018 doi: 10.29328/journal.ijcmbt.1001002; 1: 009-013
Recently Viewed
-
Obstructive Pyelonephritis Due to Postoperative Ureteral Stricture: A Case ReportAyoub Mamad*,Mohammed Amine Elafari,Mohammed Amine Bibat,Midaoui Moncef,Amine Slaoui,Tarik Karmouni,Abdelatif Koutani,Khalid Elkhader. Obstructive Pyelonephritis Due to Postoperative Ureteral Stricture: A Case Report. J Clin Med Exp Images. 2026: doi: 10.29328/journal.jcmei.1001039; 10: 003-005
-
Examining the Effects of High Poverty and Unemployment on Rural Urban Migration in Nigeria and its Consequences on Urban Resources and Rural DeclineTochukwu S Ezeudu*, Bilyaminu Tukur. Examining the Effects of High Poverty and Unemployment on Rural Urban Migration in Nigeria and its Consequences on Urban Resources and Rural Decline. J Child Adult Vaccines Immunol. 2024: doi: 10.29328/journal.jcavi.1001012; 8: 001-013
-
Case Report of a Child with Beta Thalassemia Major in a Tribal Region of IndiaNeha Chauhan, Prakash Narayan, Mahesh Narayan, Manisha Shukla*. Case Report of a Child with Beta Thalassemia Major in a Tribal Region of India. J Child Adult Vaccines Immunol. 2023: doi: 10.29328/journal.jcavi.1001011; 7: 005-007
-
Analysis and Control of a Glucose-insulin Dynamic ModelLakshmi N Sridhar*. Analysis and Control of a Glucose-insulin Dynamic Model. Ann Clin Endocrinol Metabol. 2026: doi: 10.29328/journal.acem.1001033; 10: 010-016
-
Evolution of Antifungal Activity of Artemisia herba-alba Extracts on Growth of Aspergillus sp. and Rhizopus sp.Eman MG Gebreil,Nagwa SA Alraaydi,Saleh HM EL-Majberi,Idress Hamad Attitalla*. Evolution of Antifungal Activity of Artemisia herba-alba Extracts on Growth of Aspergillus sp. and Rhizopus sp.. Int J Clin Microbiol Biochem Technol. 2025: doi: 10.29328/journal.ijcmbt.1001030; 8: 001-006
Most Viewed
-
Effects of dietary supplementation on progression to type 2 diabetes in subjects with prediabetes: a single center randomized double-blind placebo-controlled trialSathit Niramitmahapanya*,Preeyapat Chattieng,Tiersidh Nasomphan,Korbtham Sathirakul. Effects of dietary supplementation on progression to type 2 diabetes in subjects with prediabetes: a single center randomized double-blind placebo-controlled trial. Ann Clin Endocrinol Metabol. 2023 doi: 10.29328/journal.acem.1001026; 7: 00-007
-
Physical Performance in the Overweight/Obesity Children Evaluation and RehabilitationCristina Popescu, Mircea-Sebastian Șerbănescu, Gigi Calin*, Magdalena Rodica Trăistaru. Physical Performance in the Overweight/Obesity Children Evaluation and Rehabilitation. Ann Clin Endocrinol Metabol. 2024 doi: 10.29328/journal.acem.1001030; 8: 004-012
-
Hypercalcaemic Crisis Associated with Hyperthyroidism: A Rare and Challenging PresentationKarthik Baburaj*, Priya Thottiyil Nair, Abeed Hussain, Vimal MV. Hypercalcaemic Crisis Associated with Hyperthyroidism: A Rare and Challenging Presentation. Ann Clin Endocrinol Metabol. 2024 doi: 10.29328/journal.acem.1001029; 8: 001-003
-
Exceptional cancer responders: A zone-to-goDaniel Gandia,Cecilia Suárez*. Exceptional cancer responders: A zone-to-go. Arch Cancer Sci Ther. 2023 doi: 10.29328/journal.acst.1001033; 7: 001-002
-
The benefits of biochemical bone markersSek Aksaranugraha*. The benefits of biochemical bone markers. Int J Bone Marrow Res. 2020 doi: 10.29328/journal.ijbmr.1001013; 3: 027-031

If you are already a member of our network and need to keep track of any developments regarding a question you have already submitted, click "take me to my Query."