More Information
Submitted: 16 April 2020 | Approved: 27 May 2020 | Published: 28 May 2020
How to cite this article: Bakht A, Rasool N, Iftikhar S. Characterization of plastic degrading bacteria isolated from landfill sites. Int J Clin Microbiol Biochem Technol. 2020; 3: 030-035.
DOI: 10.29328/journal.ijcmbt.1001013
Copyright License: © 2020 Bakht A, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Characterization of plastic degrading bacteria isolated from landfill sites
Afreen Bakht1, Nouman Rasool2* and Saima Iftikhar3
1Department of Life Science, University of Management and Technology, Lahore, Pakistan
2Center for Professional Studies, Lahore, Pakistan
3School of Biological Sciences, University of the Punjab, Lahore, Pakistan
*Address for Correspondence: Nouman Rasool, Center for Professional Studies, Lahore, Pakistan, Email: [email protected]
The plastic pollution is threatening the environment because it has very slow degradation rate and high usage in regular activities. The present study aims at the isolation of novel microorganisms that would assist faster degradation process of polyethylene. The waste samples were collected from different landfills and dumpsites. Out of forty samples, eight samples were found to degrade polythene strips in liquid medium. Further screening of these samples showed that two strains of microbes had high potential for polythene degradation. Biochemical tests and ribotyping were performed for characterization of isolated bacteria. Resultantly, two novel bacterial strains were identified named; Bacillus wudalianchiensis_UMT (2A) and Pseudomonas aeruginosa_UMT (6). Analysis of these microbes further revealed that Bacillus wudalianchiensis_UMT and Pseudomonas aeruginosa_UMT have capability to degrade 6.6% and 4.8% polyethylene respectively. So, the results disclosed that these bacteria have great potential to degrade polythene in less time as compare to natural degradation process and can contribute to reduce pollution from our environment.
The use of plastic is growing day by day. Its durability and stability has made it an indispensable part of our life. Despite its superiority over other materials, it is posing a major threat to the environment because these long chain polymeric molecules take thousand years to decompose naturally. According to UNDP, more than 3.3 million tons of plastic are wasted in Pakistan each year. So, it is a much needed step to reduce its lifespan in order to protect the environment. Biodegradation is an environmental friendly method to degrade polyethylene as it does not generate any harmful byproducts. The speed of degradation depends upon the chemical nature of polymer, on the environmental conditions in which it is placed and on the amount of polymer. Statistically stated, 38% plastic is used for packaging and this rate of utilization is increasing 12% every year. 12% plastic is used for construction, 21% for indoors and the outdoors, 7% for automotive, 6% for electronics and 28% plastic is used in other sectors. When these plastics are introduced into the environment, they cause potential hazards [1]. Though, plastic is useful in many applications but it poses serious effect to the environment. Because plastic is durable that’s why it is highly persistent to decay. There is chemical bonding in its molecules which make is resilient to degrade. Plastic takes hundreds of years to degrade naturally [2].
Statistical data of worldwide consumption of plastic shows that North America utilizes 26%, Europe 23%, India 5%, Japan 6% and Asia contributes 40% of total plastic consumption in the world. Plastic can be disposed by different methods like incineration, landfills and through process of recycling. There are benefits and limitations in each case. When plastic waste is disposed in landfills, it apparently reduces wastes from upper surface of land but in true terms it reduces the land for agriculture [3]. As it takes so many years to degrade it naturally, so the area of landfill cannot be used for other purpose [4]. Incineration is a better option as compared to the landfills as it does not require much space and it is also better in terms of energy recovery [5]. However, it is not beyond the limitations because it causes greenhouse gases and it is also responsible for PCBs and free radical exposure [6]. Regardless of this, the limitations of landfills and incineration can be overcome through recycling though this process is expensive and the quality of end products is also low [7].
Biodegradation is the best methods as compare to the others. It is cheap and does not generate harmful pollutants into the environment. Plastic can be degraded through microorganisms. Thus, microbes play an important role in degradation process [8].
Sample collection
Forty soil samples were collected from different dumpsites and landfills from Lahore and Bhera (Pakistan) with the help of sterilized spatula and were stored in polythene bags. Each sample was given a specific ID.
Screening of microbes having ability to degrade plastic
Flasks of 5ml liquid broth were prepared. 10mg soil of each sample was put in every flask. All flasks were kept in shaker overnight to get maximum growth of microbes [9]. Took 1ml from primary inoculums and was put in 150ml enriched media. This enriched media was composed of yeast extract 0.1g, MgSO4.H2O, 0.25g; KH2PO4, 5.8g; K2HPO4, 3.7g; KNO3, 2.0g in 1000ml of distilled water. 100 mg LMWPE strips were put in each enriched media flask. These flasks were then autoclaved. After that, flasks were kept in shaker for seven days. During seven days all other microbes died except of those who had capacity to degrade polymer [10]. Agar plates were prepared from enriched media by using three different substrates to examine the efficiency of microbes in each substrate. The used substrates were consisted of 1g Polyethylene glycol (PEG), mixture of 200 µl hexadecane and 20µl Tween80 and 100 mg plastic powder [11].
Methods for screening of microbes
Clear zone method: Emulsified agar plates having substrate were streaked by dipping the L shaped loop in enriched median flasks which were previously inoculated in shaker for seven days. These plates were kept in incubator at 37ºC for seven days. After seven days, the diameter of clear zone around the colonies was observed and measured [12].
Determination of weight loss: The pre weighed plastic strips were removed from the flaks. These strips were rinsed thoroughly once with distilled water and then with absolute ethanol in order to remove the impurities. Strips were over dried at 90 ºC. The weight was determined by using the following formula [13]:
Sequencing
After isolation of genomic DNA, PCR was run for those eight samples. Universal primers 16S rRNA (5’-AAACTC/TAAAG/TGAATTGACGG-3’) and (5’-ACGGGCGGGTGTGTA/GC-3’) were used. The PCR product was analysed on 1% agarose gel and purified using gel extraction kit (Fermentas, USA). M13 forward and reverse primers were used to sequence the rRNA amplified products in a Beckman Coulter CEQTM 8000 Genetic Analyser. National Centre for Biotechnology Information BLAST database was used to amplify nucleotide sequence. These sequences were aligned with selected genera representative members by using CLUSTAL W program [14]. After sequencing, two samples (2A, 6) were preceded further which had high potential to degrade plastic more and which had new strains.
Quantification of DNA
After isolation, the genomic DNA was quantified at 260nm using UV-1280 Spectrophotometer using water as blank. Liquid media was inoculated with bacterial colonies and kept in shaker at 37ºC for overnight. After 16 hours, 1ml culture from overnight media was taken and put in 50ml media flask. The flasks were then kept in shaker and readings were taken after every two hours [15].
Effect of temperature on growth
The isolated bacterial strains were grown at temperature 37ºC, 45ºC, 50ºC and 60ºC to determine the temperature for optimal growth. After seven hours, the absorbance was assessed at 260nm by using spectrophotometer [16].
Phylogenetic tree
The sequenced 16S rRNAs amplified from both bacterial strains were compared with sequences available in nucleotide databases. Pseudomonas aeruginosa_UMT sequence was used as out group for Bacillus strains and Bacillus wudalenchinesis_UMT sequence was used as an out group for Pseudomonas aeruginosa. Best fit model was deduced and was implemented to draw phylogenetic tree.
Identification tests for microbes
Preliminary tests: Gram staining method: This is a widely used method in bacteriology. In this method, a slide was used which was grease free and clean. A smear from bacterial culture was spread in this slide by using a sterilized loop. This slide is then fixed with the heat and was passed by staining reagents [17].
Hanging drop method: This is a motility test which is used to observe live microbes and their mobility. A drop was placed on the cover slip to observe microbes in this drop and was covered by the cavity slide. Microbes in droplet were observed under light microscope [18].
Biochemical tests
Catalyze test: Culture sample was placed on the slide and then hydrogen peroxide was added on the slide drop by drop. Samples which showed bubble formation indicated positive results [19].
Motility test: The mobility test of both isolated strains was performed in semi-solid agar medium containing 0.4 g agar, 2 mg peptone and 1 g NaCl in 200ml of distilled water. The sterilized straight wire was used to inoculate the culture in a single stab centrally from top to bottom of the medium. The incubation of tube was carried out at 30ºC for overnight. A diffused hazy growth represented motile bacterium, spread throughout the medium.
Caesin hydrolysis
In this method a loop was used to transfer culture on the agar plate. Plates were inverted and kept in incubator for one to two days [20].
Gelatin hydrolysis
Suspension was prepared from overnight culture media. This suspension is then pipette in two tubes that contained gelatin agar base. Both of these tubes were kept in incubator at 37ºC. One tube was tested after 4 hours and one tube was tested after 24 hours. Acid was added and reading was recording after 30 minutes against blank gelatin agar tube.
Urea broth
In this method equal volume of urea buffer and suspension were mixed. Capillary tubes were incubated at 37 ºC and alkali production was recorded after 4 and 24 hours [21].
Oxidase test and mannitol test
The smear of a colony of isolated bacterium was introduced on a filter paper containing few drops of oxidase reagent (0.1 g of tetramethyl-p-phenylenediamine dihydrochloride in 100 ml of distilled water). The colony having oxidase activity, gave a characteristic color. In order to check the ability of isolated bacteria for fermenting mannitol, phenol red mannitol broth with 0.5-1.0% mannitol was used. Single colony of each isolated bacterium was incubated at 37 ºC for 24 hours. The change in color from red to yellow indicated that bacteria have ability to ferment mannitol.
Total forty samples were collected from different dumpsites and landfill sites of Bhera and Lahore respectively. Out of which only nine samples showed positive results i.e. reduction in weight loss (Table 1).
| Table 1: Loss of weight of plastic by isolated microbes: | ||||
| Serial No | Sample ID | Final weight | Initial weight | Weight loss (%) |
| 1 | 1A | 96.6 | 100 mg | 3.4 |
| 2 | 2A | 93.6 | 100 mg | 6.4 |
| 3 | 8A | 96.3 | 100 mg | 3.7 |
| 4 | 8B | 97.7 | 100 mg | 2.3 |
| 5 | 6 | 95.7 | 100 mg | 4.3 |
| 6 | N6 | 99 | 100 mg | 1 |
| 7 | N7(i) | 97 | 100 mg | 3 |
| 8 | N7(ii) | 98.5 | 100 mg | 1.5 |
| 9 | N9 | 99 | 100 mg | 1 |
Plate assay showing plastic degradation by microbes
Culture flasks in which weight of strips were reduced was streaked on nutrient agar plates in which PEG as substrate was added previously. After seven days of incubation, clear zones were observed around single colony having PEG substrate (Figure 1).
Figure 1: Formation of enriched zones by microbes.
Isolation of genomic DNA from microbes
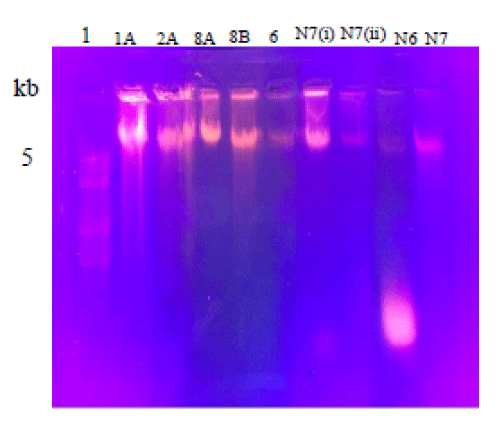
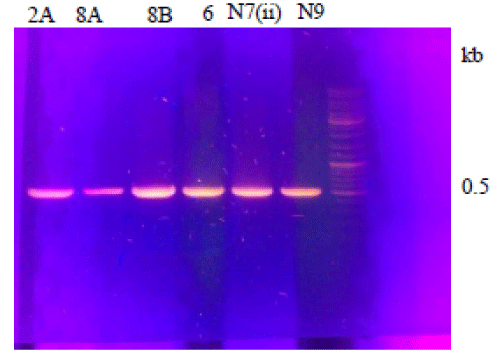
After isolation, the quality and quantity of genomic DNA was analyzed on 1% agarose gel (Figures 2,3).
Figure 2: Ethidium Bromide stainedAgarose gel showing genomic DNA isolate from different strains. Lanes 1A, 2A, 8A, 8B, 6, N7(i) showing bands of polyethylene degrading strains but very weak bands in lane N7(ii), N6 and N9. Lane 1 is showing DNA marker.
Figure 3: Ethidium Bromide stained Agarose gel showing PCR product from different strains. Lane 1 is showing ladder and lanes 2A, 8A, 8B, 6, N7(ii) and N9 showing amplified PCR product of 0.5 kb.
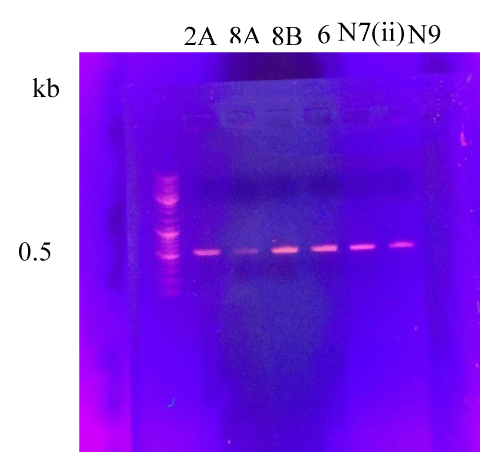
Gel of gene clean
After PCR, gene clean was done. Six samples were run for gene clean on agarose gel. After running these samples on agarose gel, positive results were seen in all samples (Figure 4).
Figure 4: Ethidium Bromide stained Agarose gel showing DNA purification gel from different samples. Lane 1 is showing ladder and lanes 2A, 8A, 8B, 6, N7(ii) and N9 showing bands of gene clean of 0.5 kb.
Ribotyping
After gene clean, ribotyping was done in which nucleotides sequences of isolated bacteria were used to analyze through NCBI BLAST database. The homology was determined by aligning the multiple sequences through CLUSTAL W program. Sequencing was done for all samples, out of which sample 2A and sample 6 had new strains.
Bacillus Sp. (Sample 2A):
CATGTGGTTTATTCGAAGCAACGCGAAGAACCTTACCAGGT CTTGACATCCCGCTGACCGGTCTGGAGACARATCTTTCCCTTCG GGGACAGCGGTGACAGGTGGTYCATGGTTGTCGTCAGCTCGTG TCGTGAGATGTTGGGTTAAGTCCCGCAACGAGCGCAACCC TTGATCTTAGTTGCCAGCATTCAGTTGGGCACTCTAAGGT GACTGCCGGTGACAAACCGGAGGAAGGTGGGGATGACGTCAAA TCATCATGCCCCTTATGACCTGGGCTACACACGTGCTACAATG GATGGTACAAAGGGCTGCAAGACCGCAAGGTTTAGCCAATCCC ATAAAACCATTCTCAGTTCGGATTGCAGGCTGCAACTCGCCTG CATGAAGCCGGAATCGCTAGTAATCGCGGATCAGCATGCCGCG GTGAATACGTTCCCGGGCCTTGWACACACCGCCCRAAA.
Pseudomonas Sp. (Sample N9):
CATGTGGTTTATTCGAGCAACGCGAAGAACCTTACCTGG CCTT\GACATGCTGAGAACTTTCCAGAGATGGATTGGKGCCTT CGGGAACTCAGACACAGGTGCTGCWTGGCTGTCGTCAGCTCG TGTCGTGAGATGTTGGGTTAAGTCCCGTAACGAGCGCAACCC TTGTCCTTAGTTACCAGCACCTCGGGTGGGCACTCTAAGGAGA CTGCCGGTGACAAACCGGAGGAAGGTGGGGATGACGTCAA GTCATCATGGCCCTTACGGCCAGGGCTACACACGTGCTACAATG GTCGGTACAAAGGGTTGCCAAGCCGCGAGGTGGAGCTAATCC CATAAAACCGATCGTAGTCCGGATCGCAGTCTGCAACTCGA CTGCGTG AAGTCGGAATCGCTAGTAATCGTGAATCAGAATGTCA CGGTGAATACGTTCCCGGGCCTTGTACACACCGCCCGT.
In vitro degradation of plastic by Pseudomonas aeruginosa_UMT and Bacillus wudalianchiensis UMT:
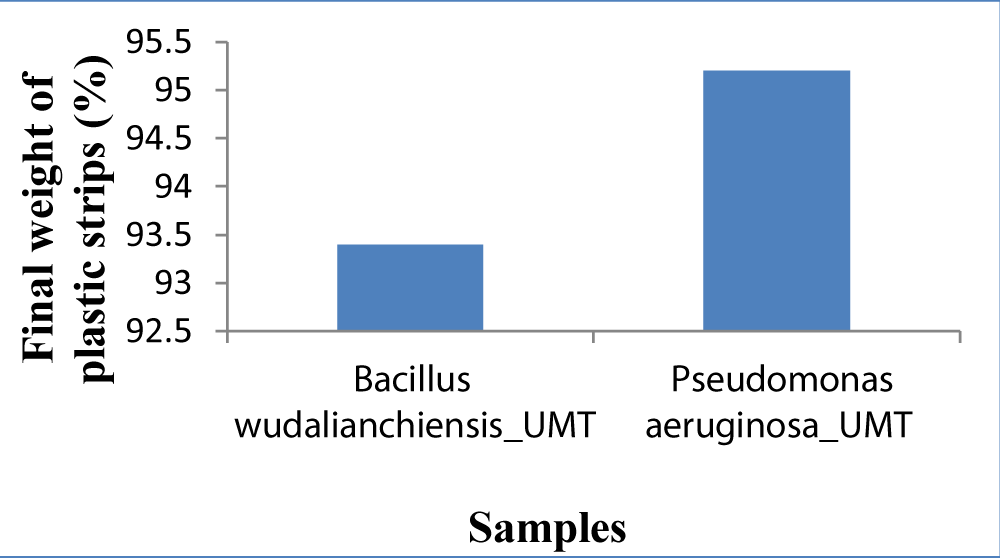
After sequencing new bacterial strains were identified that were Bacillus wudalianchiensis_UMT (2A) and Pseudomonas aeruginosa_UMT (6). The seven days experiment was repeated again that was explained previously for the confirmation of their weight reduction potential (Figure 5) (Table 2).
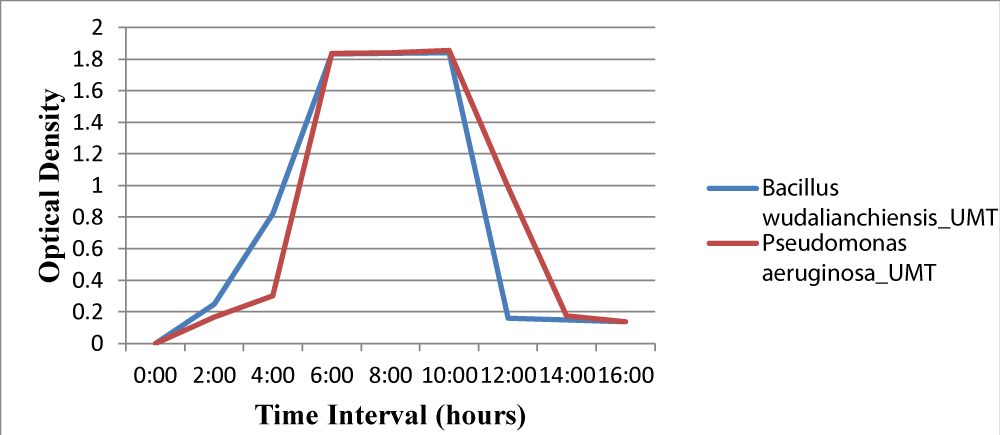
Figure 5: Graph showing comparison of final weight of both samples.
| Table 2: Percentage of weight loss by Bacillus wudalenchinesis_UMT (2A) and Pseudomonas aeruginosa_UMT (6): | ||||
| Serial No | Sample ID | Final weight (mg) |
Initial weight (mg) |
Percentage of weight lose (%) |
| 1 | 2A | 93.4 | 100 mg | 6.6 |
| 2 | 6 | 95.2 | 100 mg | 4.8 |
Effect of temperature on growth
The growth temperature of the Bacillus wudalianchiensis_UMT and Pseudomonas aeruginosa_UMT ranged from 37°C to 60°C. Growth stopped above 60°C [22] (Figure 6) (Tables 3,4).
Figure 6: Growth comparison of Bacillus wudalianchiensis_UMT and Pseudomonas aeruginosa_UMT on different time intervals at 37ºC. The growth curve indicates maximum growth at 10 hours after which it starts decreasing.
| Table 3: Determination of optimum growth of Bacillus wudalenchinesis_UMT(2A) at different temperatures. | ||
| Serial No | Temperature | Maximum OD at 260 nm |
| 1 | 37ºC | 1.839 |
| 2 | 45ºC | 0.239 |
| 3 | 50ºC | 0.342 |
| 4 | 60ºC | 0.34 |
| Table 4: Determination of optimum growth of Pseudomonas aeruginosa_UMT(6) at different temperatures | ||
| Serial No | Temperature | Maximum OD at 260 nm |
| 1 | 37ºC | 1.99 |
| 2 | 45ºC | 0.38 |
| 3 | 50ºC | 0.224 |
| 4 | 60ºC | 0.038 |
Biochemical characterization
Bacillus wudalianchiensis_UMT exhibited catalase, caesin Hydrolysis, Gelatin Hydrolysis and Urea broth activities. It indicated negative results for the tests of oxidase, mannitol and hanging drop method. Pseudomonas aeruginosa_UMT showed positive results for oxidase, mannitol, catalase, caesin hydrolysis, gelatin hydrolysis, urea broth activities and hanging drop method [23].
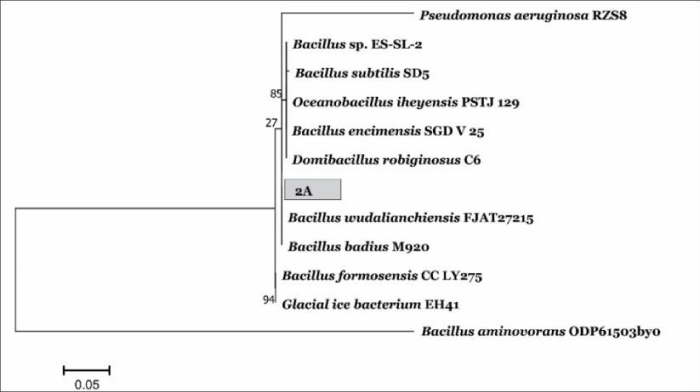
Phylogenetic tree of Bacillus wudalianchiensis_UMT: The phylogenetic tree was constructed using amplified 16S rRNA from both bacteria, sequenced and comparing with those in nucleotide databases. Pseudomonas aeruginosa_UMT was used as an out group for Bacillus wudalianchiensis_UMT. Phylogenetic tree gives a clear indication that there is homology between different strains of Pseudomonas Sp. It is clear from phylogenetic tree that sample 2A is close in cluster with Bacillus wudalianchiensis_UMT. Each name at the wudalianchiensis_UMT (Figure 7).
Figure 7: A phylogenetic tree of Bacillus_UMT (2A) showing relationships between 16S rRNA sequences.
Phylogenetic tree of Pseudomonas aeruginosa_UMT: Bacillus wudalianchiensis_UMT was used as an out-group for Pseudomonas aeruginosa. The phylogenetic tree shows that there is 90% homology between different strains of Pseudomonas (Figure 8) .
Figure 8: A phylogenetic tree of sample Pseudomonas aeruginosa_UMT (6) showing relationships between 16S rRNA sequences.
The increase in use of plastic is growing concern and major contributor to pollution across the globe. Scientists are working to develop environmental friendly methods to degrade the plastic causing pollution. Different microbes has been identified having the potential to degrade polythene and other plastics. This study is aimed at isolation and characterization of plastic degrading bacterial from landfill sites. Samples from landfill sites were used to isolate different bacteria having ability to degrade plastic. Two bacterial strains have shown great potential to degrade plastic with substantial growth in culture media. On the basis of biochemical tests and phylogenetic tree it was revealed that these two bacteria are Bacillus wudalianchiensis_UMT and Pseudomonas aeruginosa_UMT. Bacillus wudalianchiensis_UMT degraded 6.6% plastic and Pseudomonas aeruginosa_UMT degraded 4.8% in culture media. We concluded that these two bacteria are suitable to be used at large scale for degrading plastic at landfill sites and in industries to degrade plastics. In future studies, the cloning and expression of plastic degrading genes can be done.
- Kint D, Muñoz‐Guerra S. A review on the potential biodegradability of poly (ethylene terephthalate). Polymer Int. 1999; 48: 346-352. PubMed:
- Shaw DK, Sahni P. Plastic to oil. J Mechan Civil Engineering. 2004; 46-48.
- Zhang J, Wang X, Gong J, Gu Z. A study on the biodegradability of polyethylene terephthalate fiber and diethylene glycol terephthalate. J Applied Polymer Sci. 2004; 93: 1089-1096.
- Tansel B, Yildiz BS. Goal-based waste management strategy to reduce persistence of contaminants in leachate at municipal solid waste landfills. Environment, Development and Sustainability. 2001; 13: 821-831.
- Sinha V, Patel MR, Patel JV. PET waste management by chemical recycling: a review. J Polymers Environ. 2010; 18: 8-25.
- Astrup T, Møller J, Fruergaard T. Incineration and co-combustion of waste: accounting of greenhouse gases and global warming contributions. Waste Manag Res. 2009; 27: 789-799. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/19748939
- Yamada-Onodera K, Mukumoto H, Katsuyaya Y, Saiganji A, Tani Y. Degradation of polyethylene by a fungus, Penicilliumsimplicissimum YK. Polymer Degradation and Stability. 2001; 72: 323-327.
- Head IM, Swannell, RP. Bioremediation of petroleum hydrocarbon contaminants in marine habitats. Curr Opin Biotechnol. 1999; 10: 234-239. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/10361073
- Santhini K, Myla J, Sajani S, Usharani G. Screening of Micrococcus sp from oil contaminated soil with reference to bioremediation. Botany Res Int. 2009; 2: 248-252.
- Yoon MG, Jeon HJ, Kim MN. Biodegradation of polyethylene by a soil bacterium and AlkB cloned recombinant cell. J Bioremed Biodegrad. 2012; 3: 1-8.
- Kim MN, Park ST. Degradation of poly (L‐lactide) by a mesophilic bacterium. J Appl Polymer Sci. 2010; 117: 67-74.
- Augusta J, Müller RJ, Widdecke HA. A rapid evaluation plate-test for the biodegradability of plastics. Appl Microbiol Biotechnol. 1993; 39: 673-678.
- Usha R, Sangeetha T, Palaniswamy M. Screening of polyethylene degrading microorganisms from garbage soil. Libyan Agri Res Center J Int. 2011; 2: 200-204.
- Kwon SW, Kim JS, Park IC, Yoon SH, Park DH, et al. Pseudomonas koreensis sp. nov, Pseudomonas umsongensis sp. nov. and Pseudomonas jinjuensis sp. nov, novel species from farm soils in Korea. Int J Syst Evol Microbiol. 2003; 53: 21-27. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/12656147
- Rasool N, Rashid N, Javed MA, Haider MS. Requirement of pro-peptide in proper folding of subtilisin-like serine protease TK0076. Pak J Bot. 2011; 43: 2059-2065.
- Kapri A, Zaidi MGH, Satlewal A, Goel R. SPION-accelerated biodegradation of low-density polyethylene by indigenous microbial consortium. Int Biodeterio Biodegrad. 2010; 64: 238-244.
- Jalal A, Rashid N, Rasool N, Akhtar M. Gene cloning and characterization of a xylanase from a newly isolated Bacillus subtilis strain R5. J Bioscience Bioengineering. 2009; 107: 360-365.
- Vignesh R, Deepika RC, Manigandan P, Janani R. Screening ofdegrading microbes from various dumped soil samples. Int Res J Engineering Technol. 2016; 4: 2493-2498.
- Ribón W. Biochemical isolation and identification of mycobacteria. In Biochemical Testing. InTech. 2012.
- Brown A, Smith H. Benson's Microbiological Applications, Laboratory Manual in General Microbiology, Short Version. McGraw-Hill Science/Engineering/Math. 2014.
- Del Valle LJ, Jaramillo ML, Talledo M, Pons MJ, Flores L, et al. Development of a 16S rRNA PCR-RFLP assay for Bartonella identification: applicability in the identification of species involved in human infections. Uni J Microbiol Res. 2014.
- Munna MS, Tahera J, Afrad MMH, Nur IT, Noor R. Survival of Bacillus Spp. SUBB01 at high temperatures and a preliminary assessment of its ability to protect heat-stressed Escherichia coli cells. BMC Res Notes. 2015; 8: 637. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/26526722
- Amako K, Takade A. The fine structure of Bacillus subtilis revealed by the rapid-freezing and substitution-fixation method. Microscopy. J Electron Microsc (Tokyo). 1985; 34: 13-17. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/3932576